(The following article includes simple exercises in spaced repetition using Orbit, an interesting system for learning under development by Andy Matuschak. If you register at the Orbit site, you will be periodically emailed an opportunity to review information in this article. The idea, as Andy says, is to improve learning and retention by extending the reading experience over time via a mnemonic medium).
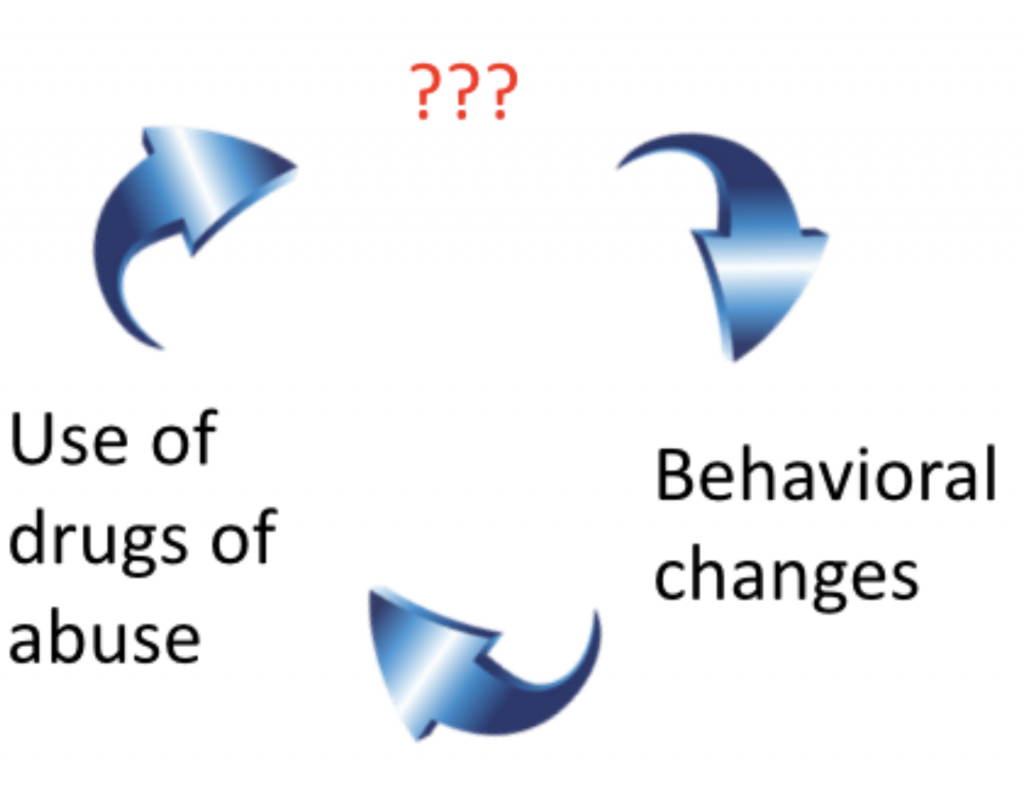
We humans think we have free will, but find ourselves foiled at every turn
Our biology conspires against us with brains hardwired to increase pleasure and decrease pain.
Nora Volkow
“Doctor, why can’t I stop using drugs?”
Consider an all-too-frequently-reported history of drug exposure.You are reviewing substance use history with your new patient, a 44 year-old man:
- Starting smoking cigarettes age 13, still smokes tobacco.
- Huffed paint and nitrous oxide around age 13.
- First alcohol drink age 14, blackouts x 9, still uses alcohol “to come down”
- Started smoking cannabis age 14, use persists.
- First use of cocaine age 19, used x 4 years heavily “in the 80s”.
- First use of methamphetamine 12 years ago, used daily last six years.
- He has had enormous adverse life impacts from use of these substances: loss of jobs, incarceration, loss of relationships.
What’s happened to the locus of control (the brain) that could explain ongoing use despite substantial risk?
Stimulants, nicotine, opioids, alcohol all increase levels of dopamine (DA) in the nucleus accumbens. Food, sex, and other activities also cause increased levels there, but at much lesser levels. This is the classic picture of the centrality of dopamine in reward and reinforcement leading to drug addiction.
But that doesn’t tell a complete story.
A more detailed picture comes from imaging studies done with radiolabelled raclopride, a dopamine (DA) D2 antagonist. Binding of this PET ligand is inversely related to DA levels in regions of interest.
One prediction we might make is that DA levels in the striatum will be correlated with euphoric feelings in the user – the “high” of the drug experience. However, Dr. Nida Volkow’s studies (NIH/NIDA) show that it is the speed at which a drug enters the brain (not the quantity) that corresponds to more DA availability and more of a “high”. This observation matches what we see in stimulant-using subjects, who generally associate a”high” more with smoking or intravenous use when compared to oral use.
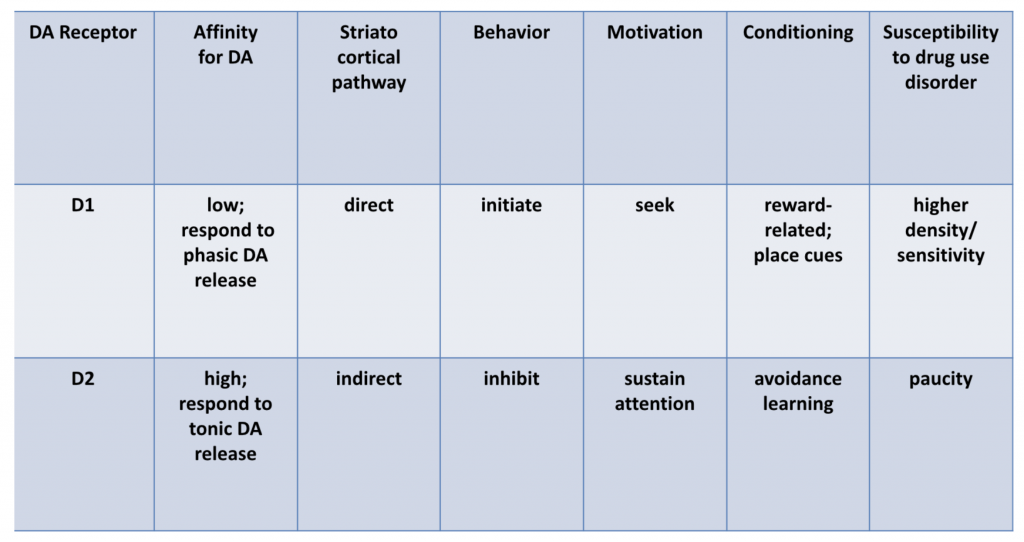
Dopamine: the importance of D1, D2, and tonic and phasic activity
One neurotransmitter to ring them,
One neurotransmitter to bring them,
One neurotransmitter to find them
and in the darkness bind them.
In the Land of Addiction where the Shadows fall.
One neurotransmitter to rule them all
(apologies to J. R. R. Tolkien)
Dopamine activity falls into two general categories:
- tonic DA activity, which results in consistent but low to moderate levels of DA
- phasic DA activity, which results in frequent bursts of DA
Tonic and phasic DA activity is associated with different effects:
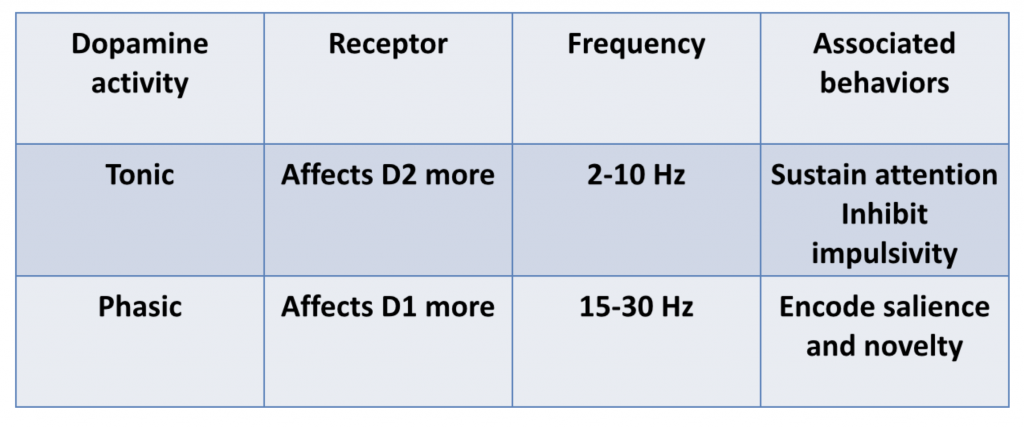
Dr. Volkow explained that D2 pathways are primarily mediated by tonic DA activity, but that phasic DA activity is needed to stimulate D1 pathways, as D1 has a lower affinity for DA than D2 receptors.
And it is precisely phasic activity that results when drugs of abuse are used, particularly when they are smoked or used intravenously.
(Indeed, this may explain why our “substitution therapies” haven’t worked in stimulant dependence. Many clinicians have asked why we don’t maintain methamphetamine abusing patients on methylphenidate or amphetamines, just as we substitute methadone or buprenorphine for heroin. In fact, prescription stimulants and novel stimulants like modafinil have all been tried as agents of harm-reduction or in hopes of reducing craving and relapse. None have been very successful in decreasing street drug use. One reason for inefficacy may be that the experience of using an oral formulation of these prescription stimulants does not result in a phasic release of DA).
Dr. Volkow goes on to note the central role D1 activity plays in impulsivity, in conditioning, and in phenomenon like the association of place cues with drug experiences. Methamphetamine-using patients often say that the sight of drug use apparatus like meth pipes or syringes, or proximity to a place of past meth purchase engenders intense drug craving. Sometimes meth users report actually salivating for the drug when in a place they’ve previously used meth.
This makes sense in terms of adaptive behavior: we experience a novel reward and receive immediate pleasurable feedback. Novel pleasurable experiences activate DA pathways and glutamatergic activity that yields more synaptic connections in reward pathways and, importantly, in the hippocampus: we have learned and will remember a salient behavior that provides an intense reward.
Nature is efficient: with such place-cue and other conditioning we no longer need to repeatedly taste food (or a drug) to be motivated to consume it – sight, sound, or scent will cue and motivate us to seek and consume. Indeed, Dr. Volkow has called such conditioning “the essence of addiction.”
D2 activity, conversely, is associated in general terms with inhibition of impulsivity. There is much interesting research showing that polymorphisms or decreased density of D2 receptor predict impulsivity and addiction [1]. Decreased D2 activity is primarily associated with reduced metabolic activity in the prefrontal cortex. Individuals with a strong family history of alcoholism show reductions in D2 receptor density.
Treatment: what can we do?
Multiple circuits (e.g., for impulse control, memory, and cognition) are disrupted in the drug use disorder brain – not just the “reward pathways.”
Dr. Volkow paints addiction as akin to an animal starving – the drug is perceived at some level as needed for survival, and drug-using behavior must therefore persist despite almost any level of risk.
In other words, the prefrontal cortex is overwhelmed (research shows demonstrable anatomic changes in the prefrontal cortex from drug exposure, as the following figure illustrates):
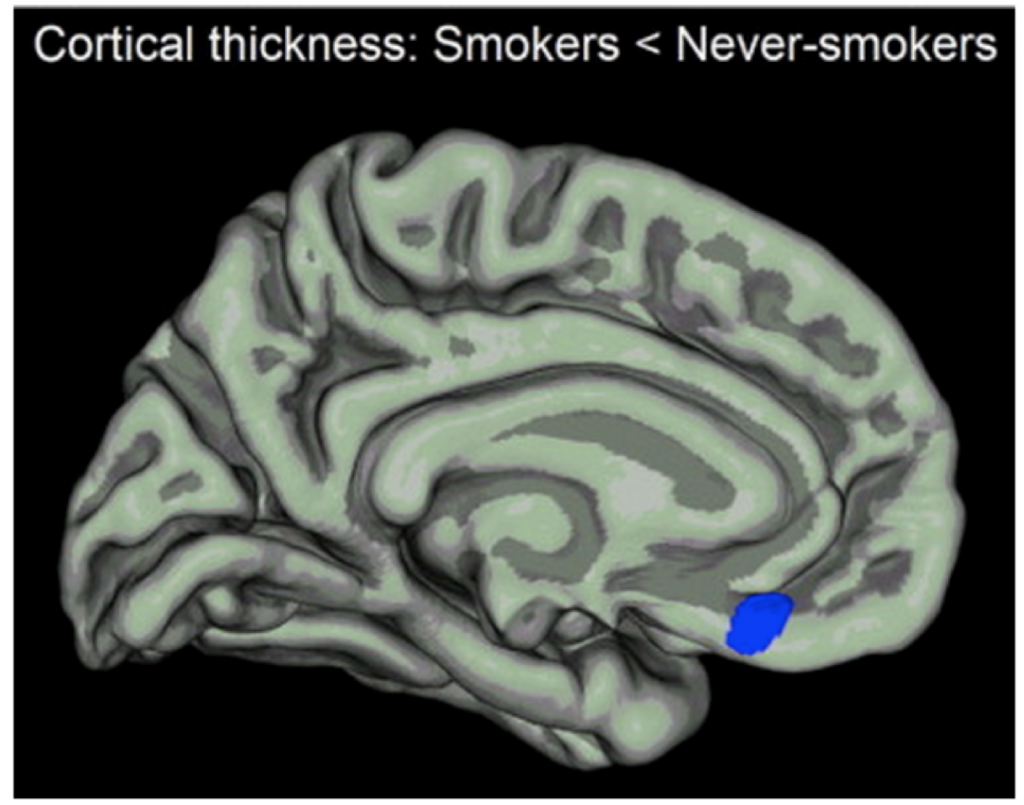
We cannot expect medication to balance or rewire all the changes that have led to such behaviors. What may be lacking are interventions that strengthen executive function and inhibit the responses that lead to continued use. Can’t we somehow attenuate the reward circuitry or otherwise resist urges (ie by enhancing cognitive pathways) that may be maladaptive?
Dr. Volkow wishes for a D2 receptor increasing agent in humans – an agent to attenuate impulsivity. None are available for clinical use, but she cites an animal study of an adenovirus carrying a D2 receptor gene into the nucleus accumbens, which subsequently reduced compulsive alcohol self-administration in animals receiving the virus.
Another treatment approach is immunotherapy to decrease the ability of drug to get into the brain.
Stress reactivity is a strong trigger and predictor of relapse, so buffering stress through medications or therapy can be useful.
Dr. Volkow advises clinicians to work with patients around avoiding cues, and to prepare patients beforehand for the triggering that follows the cue. Patients of course cannot avoid all cues. We can coach inhibition of the vental-striatal region by practicing activating the cortex in imagined triggering situations.
Alternative strategies might include the monitoring stress and even proximity to place cues as triggers (“ecological momentary assessment”), with real-time interventions. There is a role here for smartphone monitoring and intervention/reminder systems [2-5]. For example, geographical location could trigger warnings about impending place cues, and urge users to stay away. Software can periodically query for the presence of stressors and other triggers or by noting behaviors like isolation or changes in activity and sleep. Reminders can take the form of text messages, recorded audio or video, or notification and intervention by a clinician.
References
- Blum K, Sheridan PJ, Wood RC, Braverman ER, Chen TJ, Comings DE. Dopamine D2 receptor gene variants: association and linkage studies in impulsive-addictive-compulsive behaviour. Pharmacogenetics. 1995 Jun;5(3):121-41.
- See for the Android platform: “Real time tracking of symptoms for research and clinical support”;В http://apps.keithflower.org/?p=361
- Stone, AA. Shiffman, S. Ecological momentary assessment (EMA) in behavorial medicine. Annals of Behavioral Medicine, Vol 16(3), 1994, 199-202.
- Moskowitz, DS. Young SN. Ecological momentary assessment: what it is and why it is a method of the future in clinical psychopharmacology. J Psychiatry Neurosci. 2006 January; 31(1): 13–20.
- Collins RL, Kashdan TB, Gollnisch G. The feasibility of using cellular phones to collect ecological momentary assessment data: application to alcohol consumption. Exp Clin Psychopharmacol. 2003;11(1):73–78.
- Freedman MJ, Lester KM, McNamara C, Milby JB, Schumacher JE. Cell phones for ecological momentary assessment with cocaine-addicted homeless patients in treatment. J Subst Abuse Treat. 2006;30(2):105–111.